Make Multi-Wavelength Light Curves for Large Samples#
Learning Goals#
By the end of this tutorial, you will be able to:
Parallelize the code demonstrated in the light_curve_collector notebook to get multi-wavelength light curves.
Launch a run using a large sample of objects, monitor the run’s progress automatically, and understand its resource usage (CPU and RAM).
Understand some general challenges and requirements when scaling up code.
Introduction#
This notebook shows how to collect multi-wavelength light curves for a large sample of target objects. This is a scaling-up of the light_curve_collector and assumes you are familiar with the content of that notebook.
Notebook sections are:
Overview: Describes functionality of the included bash script and python helper functions. Compares some parallel processing options. Introduces
topand what be aware of.Example 1: Shows how to launch a large-scale run using the bash script, monitor its progress automatically, and diagnose a problem (out of RAM). This method is recommended for sample sizes greater than a few hundred.
Example 2: Shows how to parallelize the example from the light_curve_collector notebook using the helper and python’s
multiprocessinglibrary.Example 3: Details the helper parameter options and how to use them in python and bash.
Appendix: Contains background information including a discussion of the challenges, needs, and wants encountered when scaling up this code, and general advice for the user which can be applied to other use cases.
As written, this notebook is expected to require at least 1 CPU and 6G RAM.
Many of the bash commands below are shown in non-executable cells because they are not intended to be run in this notebook.
Bash commands that are not executed below begin with the symbol $ , and those that are executed begin with !.
Both types can be called from the command-line – open a new terminal and copy/paste the cell text without the beginning symbol. The bash script is not intended to be executed from within a notebook and may behave strangely if attempted.
Also be aware that the script path shown in the commands below assumes you are in the same directory as this notebook. Adjust it if needed.
Parallel processing methods: bash script vs. python’s multiprocessing#
Bash script: Recommended for most runs with medium to large sample sizes (>~500). Allows ZTF to use additional parallelization internally, and so is often faster (ZTF often takes the longest and returns the most data for AGN-like samples). Writes stdout and stderr to log files, useful for monitoring jobs and resource usage. Can save
topoutput to a file to help identify CPU and RAM usage/needs.Python’s
multiprocessinglibrary: Can be convenient for runs with small to medium sample sizes, up to ~500. Has drawbacks that may be significant including the inability to use ZTF’s internal parallelization and that it does not save the log output (stdout and stderr) to file. An advantage of themultiprocessingexample in this notebook over the light_curve_collector is that it automatically saves the sample and light curve data to disk after loading them and can automatically skip those functions in subsequent calls and use the files instead.
The python helper#
The python “helper” is a set of wrapper functions around the same ‘code_src/’ functions used in the light_curve_collector notebook.
The wrappers facilitate parallelization and large-scale runs by automating tasks like saving the function outputs to files.
The helper does not actually implement parallelization and can only run one function per call.
The helper can be used in combination with any parallel processing method.
The helper can load
topoutput from a file to pandas DataFrames and make some figures.
The bash script#
The bash script allows the user to collect light curves from a large scale sample with a single command and provides options to help manage and monitor the run. In a nutshell, the script does the following when called using flags that “launch a run”:
calls the helper to gather the requested sample and then launches jobs for each archive query in separate, parallel processes.
redirects stdout and stderr to log files.
tells the user what the process PIDs are and where the log and data files are.
exits, leaving the archive jobs running in the background.
In case all the archive calls need to be canceled after the script exits, the user can call the script again with a different flag to have it find and kill all the processes it launched.
While the jobs are running, the user can call the script again with a different flag to have it save top output to a log file at a user-defined interval.
The helper can be used to load this file to pandas DataFrames and make some figures.
Interpreting top#
If you are unfamiliar with top, the answer to this StackExchange question contains a basic description of the fields in the output.
Also, beware that top can be configured to display values in different ways (e.g., as a percentage of either a single CPU or of all available CPUs).
To understand the local configuration, read the man page (run man top in a terminal), particularly the sections “SUMMARY Display” and “FIELDS / Columns Display”.
Fornax Science Console#
There are a couple of things to be aware of when running on the Fornax Science Console.
Take note of the amount of CPU and RAM available to the server type you choose when starting the session.
In particular, beware that top can show a larger amount of total RAM than is actually accessible to your server due to resource sharing between users.
The Science Console is primarily intended for interactive use and will cull sessions which appear to be inactive.
If you want a notebook or script to run for longer than about 30 minutes and you will not be interacting with the Console, running top during that time can help keep the session active.
Imports#
This cell will install the dependencies, if needed:
# Uncomment the next line to install dependencies if needed.
# %pip install -r requirements_scale_up.txt
import multiprocessing # python parallelization method
import pandas as pd # use a DataFrame to work with light-curve and other data
import sys # add code directories to the path
sys.path.append("code_src/")
import helpers.scale_up # python "helper" for parallelization and large scale runs
import helpers.top # load `top` output to DataFrames and make figures
from data_structures import MultiIndexDFObject # load light curve data as a MultiIndexDFObject
from plot_functions import create_figures # make light curve figures
1. Multi-wavelength light curves for 500,000 SDSS AGN#
This example shows how to launch a large-scale collection of light curves using the bash script, monitor its performance, and diagnose a problem (out of RAM). This run collects light curves for 500,000 SDSS objects and takes several hours to complete, but is not actually executed here. Instead, we show the bash commands and then look at logs that were generated by running the commands on 2024/03/01. If executed, the run is expected to require at least 2 CPUs and 100G RAM.
1.1 Launch the run#
# This will launch a very large scale run.
# See Example 3 for a detailed explanation of parameter options.
# If you are running this for the first time, reduce the sample size 500000 -> 50.
# run_id will be used in future calls to manage this run, and also determines the name of the output directory.
# If you are executing this, change run_id to your own value to avoid overwriting the demo logs.
$ run_id="demo-SDSS-500k"
# Execute the run.
# This will get the sample and then call all archives specified by the '-a' flag.
# Defaults will be used for all keyword arguments that are not specified.
$ bash code_src/helpers/scale_up.sh \
-r "$run_id" \
-j '{"get_samples": {"SDSS": {"num": 500000}}, "archives": {"ZTF": {"nworkers": 8}}}' \
-a "Gaia HEASARC IceCube WISE ZTF"
The script will run the ‘get sample’ job, then launch the archive query jobs in parallel and exit. Archive jobs will continue running in the background until they either complete or encounter an error. Example 2 shows how to load the data.
Command output from 2024/03/01 logs:
!cat output/lightcurves-demo-SDSS-500k/logs/scale_up.sh.log
# There is a warning that the SDSS 'specObjID' column was converted from an int to a string.
# The column is immediately dropped. It is not used in the code.
*********************************************************************
** Run starting. **
run_id=SDSS-500k
base_dir=/home/jovyan/raen/fornax-demo-notebooks/light_curves/output/lightcurves-SDSS-500k
logs_dir=/home/jovyan/raen/fornax-demo-notebooks/light_curves/output/lightcurves-SDSS-500k/logs
parquet_dir=/home/jovyan/raen/fornax-demo-notebooks/light_curves/output/lightcurves-SDSS-500k/lightcurves.parquet
** **
Build sample is starting. logfile=get_sample.log
Build sample is done. Printing the log for convenience:
-- /home/jovyan/raen/fornax-demo-notebooks/light_curves/output/lightcurves-SDSS-500k/logs/get_sample.log:
2024/03/01 20:31:27 UTC | [pid=3289] Starting build=sample
Building object sample from: ['sdss']
WARNING: OverflowError converting to IntType in column specObjID, reverting to String. [astropy.io.ascii.fastbasic]
SDSS Quasar: 500000
Object sample size, after duplicates removal: 498877
Object sample saved to: /home/jovyan/raen/fornax-demo-notebooks/light_curves/output/lightcurves-SDSS-500k/object_sample.ecsv
2024/03/01 20:36:59 UTC
--
Archive calls are starting.
[pid=3968] Gaia started. logfile=gaia.log
[pid=3972] HEASARC started. logfile=heasarc.log
[pid=3976] IceCube started. logfile=icecube.log
[pid=3980] WISE started. logfile=wise.log
[pid=3984] ZTF started. logfile=ztf.log
** **
** Main process exiting. **
** Jobs may continue running in the background. **
**********************************************************************
1.2 Cancel#
You can cancel jobs at any time.
If the script is still running, press Control-C.
If the script has exited, there are two options.
To cancel an individual job, kill the job’s process using:
$ pid=0000 # get the number from script output
$ kill $pid
To cancel the entire run, use the
-k(kill) flag:
$ bash code_src/helpers/scale_up.sh -r "$run_id" -k
1.3 Restart#
If you want to restart and skip step(s) that previously completed, run the first command again and add one or both “overwrite” flags set to false:
# use the same run_id as before
$ bash code_src/helpers/scale_up.sh \
-r "$run_id" \
-j '{"get_samples": {"SDSS": {"num": 500000}}, "archives": {"ZTF": {"nworkers": 8}}}' \
-a "Gaia HEASARC IceCube WISE ZTF" \
-d "overwrite_existing_sample=false" \
-d "overwrite_existing_lightcurves=false"
1.4 Monitor#
There are at least three places to look for information about a run’s status.
Check the logs for job status or errors. The bash script will redirect stdout and stderr to log files and print out the paths for you.
Check for light curve (parquet) data. The script will print out the “parquet_dir”.
lsthis directory. You will see a subdirectory for each archive call that has completed successfully, assuming it retrieved data for the sample.Watch
top. The script will print the job PIDs. The script can also monitortopfor you and save the output to a log file.
1.4.1 Logs#
# Gaia log from 2024/03/01 (success)
!cat output/lightcurves-demo-SDSS-500k/logs/gaia.log
2024/03/01 20:37:00 UTC | [pid=3968] Starting build=lightcurves, archive=gaia
INFO: Query finished. [astroquery.utils.tap.core]
Light curves saved to:
parquet_dir=/home/jovyan/raen/fornax-demo-notebooks/light_curves/output/lightcurves-SDSS-500k/lightcurves.parquet
file=archive=gaia/part0.snappy.parquet
2024/03/01 21:07:38 UTC
# ZTF log from 2024/03/01 (failure)
!cat output/lightcurves-demo-SDSS-500k/logs/ztf.log
2024/03/01 20:37:00 UTC | [pid=3984] Starting build=lightcurves, archive=ztf
100%|███████████████████████████████████████████████████████████████████████████████| 50/50 [44:40<00:00, 53.61s/it]
2024/03/01 21:21:48 UTC | [pid=13387] Starting worker
2024/03/01 21:21:48 UTC | [pid=13388] Starting worker
2024/03/01 21:21:48 UTC | [pid=13389] Starting worker
2024/03/01 21:21:48 UTC | [pid=13390] Starting worker
2024/03/01 21:21:48 UTC | [pid=13391] Starting worker
2024/03/01 21:21:48 UTC | [pid=13392] Starting worker
2024/03/01 21:21:48 UTC | [pid=13393] Starting worker
2024/03/01 21:21:48 UTC | [pid=13394] Starting worker
100%|███████████████████████████████████████████████████████████████████████| 72397/72397 [1:38:37<00:00, 12.23it/s]
The logs above show that the ZTF job loaded the light curve data successfully (“100%”) but exited without writing the parquet file (no “Light curves saved” message like Gaia).
The data was lost.
There is also no indication of an error; the job just ends.
We can diagnose what happened by looking at the top output.
1.4.2 top#
While the jobs are running, you can monitor top and save the output to a log file by calling the script again with the -t flag.
This call must be separate from the call that launches the run.
If you want to capture the sample step as well as the archive queries, open a second terminal and call the script with -t right after launching the run in the first terminal.
$ interval=10m # choose your interval
$ bash code_src/helpers/scale_up.sh -r "run_id" -t "$interval"
The script will continue running until after all of run_id’s jobs have completed.
You can cancel at anytime with Control-C and start it again with a new interval.
Once saved to file, the helper can parse the top output into pandas DataFrames and make some figures.
top output from 2024/03/01:
run_id = "demo-SDSS-500k"
logs_dir = helpers.scale_up.run(build="logs_dir", run_id=run_id)
toplog = helpers.top.load_top_output(toptxt_dir=logs_dir, run_id=run_id)
# System summary information from lines 1-5 of `top` output. One row per time step.
toplog.summary_df.sample(5)
| load_avg_per_1min | load_avg_per_5min | load_avg_per_15min | total_GiB | free_GiB | used_GiB | avail_GiB | delta_time | |
|---|---|---|---|---|---|---|---|---|
| time | ||||||||
| 2024-03-01 23:06:08+00:00 | 1.57 | 1.93 | 2.17 | 61.8 | 34.3 | 13.9 | 47.2 | 0 days 02:34:40 |
| 2024-03-01 20:41:30+00:00 | 1.80 | 1.52 | 0.87 | 61.8 | 50.8 | 2.6 | 58.5 | 0 days 00:10:02 |
| 2024-03-01 23:03:23+00:00 | 1.81 | 2.17 | 2.28 | 61.8 | 34.3 | 13.9 | 47.2 | 0 days 02:31:55 |
| 2024-03-01 23:01:08+00:00 | 2.01 | 2.39 | 2.35 | 61.8 | 28.4 | 21.4 | 39.7 | 0 days 02:29:40 |
| 2024-03-01 21:31:41+00:00 | 2.23 | 2.25 | 1.55 | 61.8 | 40.5 | 12.4 | 49.4 | 0 days 01:00:13 |
# Information about running processes from lines 6+ of `top` output. One row per process, per time step.
toplog.pids_df.sample(5)
| job_name | pid_name | PID | USER | PR | NI | VIRTg | RESg | SHRg | S | %CPU | %MEM | TIME+ | COMMAND | delta_time | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| time | |||||||||||||||
| 2024-03-01 21:22:33+00:00 | ztf | ztf | 3984 | jovyan | 20 | 0 | 4.2 | 1.4 | 0.1 | S | 0.0 | 2.2 | 0:57.66 | python | 0 days 00:51:05 |
| 2024-03-01 23:05:05+00:00 | ztf | ztf | 3984 | jovyan | 20 | 0 | 37.9 | 9.9 | 0.1 | R | 100.0 | 16.0 | 6:05.89 | python | 0 days 02:33:37 |
| 2024-03-01 21:23:19+00:00 | ztf | ztf-worker | 13392 | jovyan | 20 | 0 | 4.3 | 1.2 | 0.0 | S | 0.0 | 1.9 | 0:18.42 | python | 0 days 00:51:51 |
| 2024-03-01 22:36:56+00:00 | ztf | ztf-worker | 13394 | jovyan | 20 | 0 | 11.2 | 2.1 | 0.0 | S | 0.0 | 3.4 | 15:08.46 | python | 0 days 02:05:28 |
| 2024-03-01 23:00:01+00:00 | ztf | ztf-worker | 13394 | jovyan | 20 | 0 | 19.4 | 1.5 | 0.0 | S | 43.8 | 2.4 | 20:59.96 | python | 0 days 02:28:33 |
fig = toplog.plot_overview()
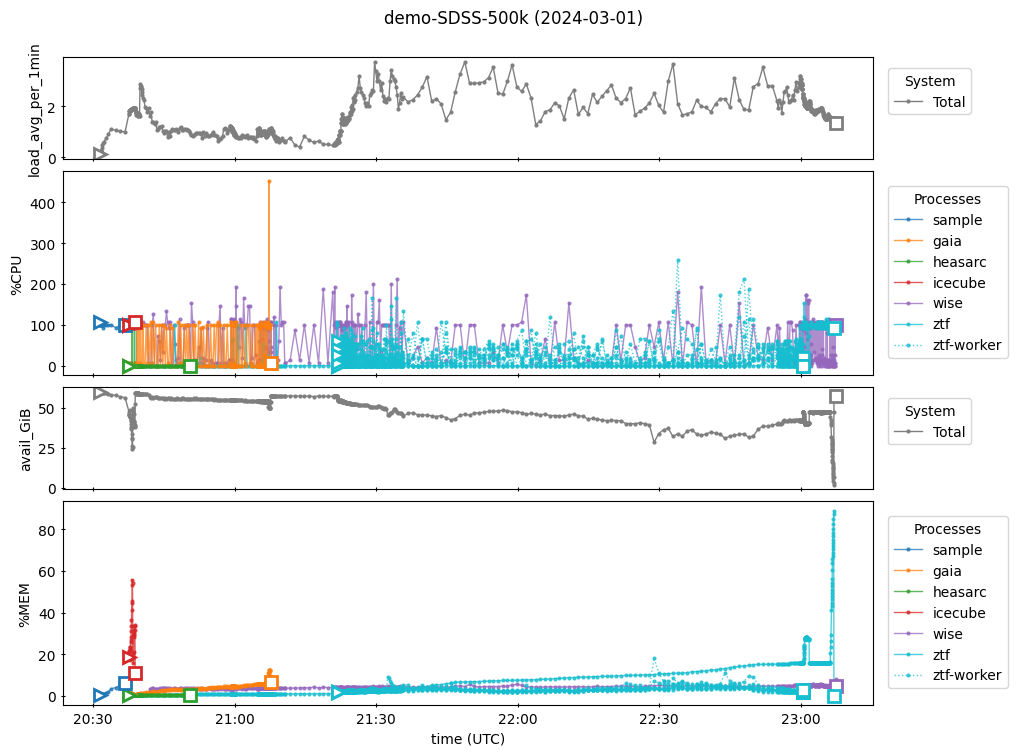
In the figure, panels 1 and 2 show CPU usage while 3 and 4 show memory usage. Panels 1 and 3 show system totals while panels 2 and 4 show usage by individual processes. Specifically:
Panel 1: System CPU load, averaged over the previous one minute. A value of “1” means that all compute tasks could have been completed by one CPU working 100% of the time (in reality, the tasks would have been split between the available CPUs). If this value exceeds the number of CPUs available, some tasks will wait in a queue before being executed.
Panel 2: Percentage of time the given process used a CPU. This can be greater than 100% if a process uses multiple threads.
Panel 3: Amount of unused memory available to the system. If this nears zero, the system will start killing processes so that the entire machine doesn’t go down.
Panel 4: Percentage of total RAM used by the given process.
There are many interesting features in the figure that the reader may want to look at in more detail. For example, other than ZTF’s memory spike at the end, we see that the full run collecting multi-wavelength light curves for the SDSS 500k sample could be completed with about 2 CPU and 60G RAM. We also see that the IceCube call completes very quickly but requires a significant amount of memory. Another observation is that the WISE call’s CPU activity seems to slow down when the ZTF workers are running, indicating that a different number of ZTF workers may be more efficient overall.
We want to learn why the ZTF job failed. Let’s zoom in on that time period:
fig = toplog.plot_overview(between_time=("22:55", "23:10"))
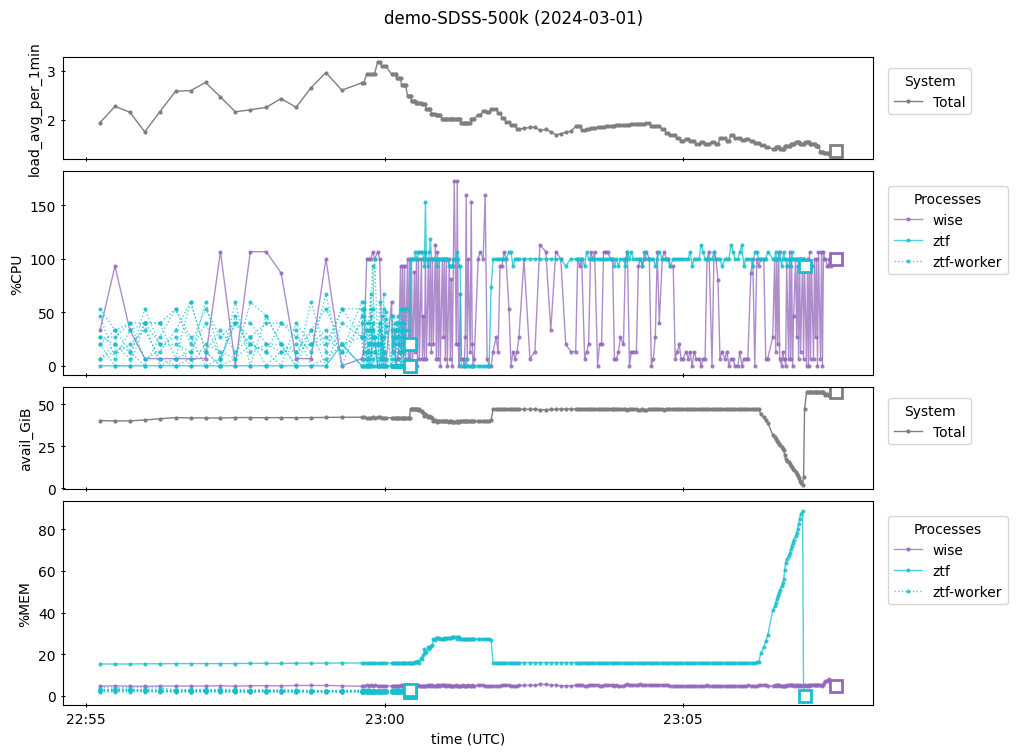
In the second panel above we see the ZTF worker processes (which load the light curve data) ending just after 23:00 and then the ZTF parent process continues by itself.
Around 23:06 in the fourth panel, we see the ZTF job’s memory usage rise sharply to almost 100% and then drop immediately to zero when the job terminates.
This coincides with the total available memory dropping to near zero in the third panel.
This shows that the machine did not have enough memory for the ZTF call to successfully transform the light curve data collected by the workers into a MultiIndexDFObject and write it as a parquet file, so the machine killed the job.
The solution is to rerun the ZTF archive call on a machine with more RAM.
To learn exactly which step in ztf_get_lightcurves was causing this and how much memory it actually needed, additional print statements were inserted into the code similar to the following:
# this was inserted just before "exploding" the dataframe in ztf_functions.py
# helpers.scale_up._now() prints the current timestamp
print(f"{helpers.scale_up._now()} | starting explode", flush=True)
# ztf_df = ztf_df.explode(... # this is the next line in ztf_functions.py
2025/09/12 01:49:54 UTC | starting explode
ZTF was then rerun on a large machine and top output was saved.
After the run, we manually compared timestamps between the ZTF log and top output and tagged relevant top timestamps with corresponding step names by appending the name to the ‘—-’ delineator, like this for the “explode” step:
!cat output/lightcurves-demo-SDSS-500k/logs/top.tag-ztf.txt | grep -A12 explode
----explode
2024/02/25 02:09:32 UTC
top - 02:09:32 up 1:20, 0 users, load average: 1.00, 0.68, 0.43
Tasks: 1 total, 1 running, 0 sleeping, 0 stopped, 0 zombie
%Cpu(s): 0.6 us, 0.2 sy, 0.0 ni, 99.2 id, 0.0 wa, 0.0 hi, 0.0 si, 0.0 st
GiB Mem : 496.0 total, 454.7 free, 13.6 used, 27.7 buff/cache
GiB Swap: 0.0 total, 0.0 free, 0.0 used. 479.4 avail Mem
PID USER PR NI VIRT RES SHR S %CPU %MEM TIME+ COMMAND
1418 jovyan 20 0 82.5g 11.8g 0.1g R 100.0 2.4 4:20.44 ipython
----
2024/02/25 02:09:34 UTC
top - 02:09:34 up 1:20, 0 users, load average: 1.08, 0.70, 0.44
The helper can recognize these tags and show them on a figure:
ztf_toplog = helpers.top.load_top_output(toptxt_file="top.tag-ztf.txt", toptxt_dir=logs_dir, run_id=run_id)
fig = ztf_toplog.plot_time_tags(summary_y="used_GiB")
# (This run starts by reading a previously cached parquet file containing the raw data returned by workers.)
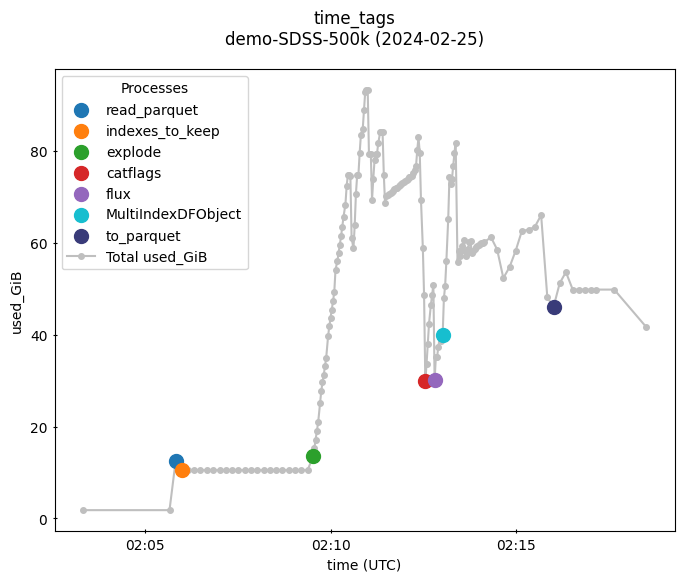
This figure shows that almost 100G RAM is required for the ZTF job to succeed.
It further shows that the “explode” step requires the most memory, followed by creating the MultiIndexDFObject.
From here, the user can choose an appropriately sized machine and/or consider whether ztf_get_lightcurves could be made to use less memory.
2. Parallelizing the light_curve_collector notebook#
This example shows how to parallelize the example from the light_curve_collector notebook using the helper and python’s multiprocessing.
An advantage of the method shown here is that it automatically saves the sample and light curve data to disk after loading them and can automatically skip those steps in subsequent calls and use the files instead.
This is a small sample (Yang, 30 objects).
If you want a sample larger than a few hundred, consider using the bash script instead.
Define the keyword arguments for the run:
kwargs_dict = {
"run_id": "demo-Yang-sample",
# Paper names to gather the sample from.
"get_samples": ["Yang"],
# Keyword arguments for *_get_lightcurves archive calls.
"archives": {
"Gaia": {"search_radius": 1 / 3600, "verbose": 0},
"HEASARC": {"catalog_error_radii": {"FERMIGTRIG": 1.0, "SAXGRBMGRB": 3.0}},
"IceCube": {"icecube_select_topN": 3, "max_search_radius": 2.0},
"WISE": {"radius": 1.0, "bandlist": ["W1", "W2"]},
"ZTF": {"match_radius": 1 / 3600, "nworkers": None},
},
}
# See Example 3 for a detailed explanation of parameter options.
kwargs_dict
{'run_id': 'demo-Yang-sample',
'get_samples': ['Yang'],
'archives': {'Gaia': {'search_radius': 0.0002777777777777778, 'verbose': 0},
'HEASARC': {'catalog_error_radii': {'FERMIGTRIG': 1.0, 'SAXGRBMGRB': 3.0}},
'IceCube': {'icecube_select_topN': 3, 'max_search_radius': 2.0},
'WISE': {'radius': 1.0, 'bandlist': ['W1', 'W2']},
'ZTF': {'match_radius': 0.0002777777777777778, 'nworkers': None}}}
Decide which archives to query. This is a separate list because the helper can only run one archive call at a time. We will iterate over this list and launch each job separately.
# archive_names = ["PanSTARRS", "WISE"] # choose your own list
archive_names = helpers.scale_up.ARCHIVE_NAMES["all"] # predefined list
archive_names
['Gaia',
'HCV',
'HEASARC',
'IceCube',
'PanSTARRS',
'TESS_Kepler',
'WISE',
'ZTF']
In the next cell, we:
collect the sample and write it as a .ecsv file; then
query the archives in parallel using a
multiprocessing.Pooland write the light curve data as .parquet files.
Note: Since the next cell launches each archive call in a child process (through multiprocessing.Pool),
the archive calls themselves will not be allowed to launch child processes of their own.
We’ll just skip the affected archive calls here and encourage the user towards the bash script since this
is one of the main reasons that it was developed – it launches the archive jobs in parallel, but in a
different way, so that the calls are also allowed to parallelize their own internal code and thus run much faster.
%%time
# Skip archive calls that use internal parallelization.
_archive_names = [name for name in archive_names if name.lower() not in ["panstarrs", "ztf"]]
sample_table = helpers.scale_up.run(build="sample", **kwargs_dict)
# sample_table is returned if you want to look at it but it is not used below
with multiprocessing.Pool(processes=len(_archive_names)) as pool:
# submit one job per archive
for archive in _archive_names:
pool.apply_async(helpers.scale_up.run, kwds={"build": "lightcurves", "archive": archive, **kwargs_dict})
pool.close() # signal that no more jobs will be submitted to the pool
pool.join() # wait for all jobs to complete
# Note: The console output from different archive calls gets jumbled together below.
# Worse, error messages tend to get lost in the background and never displayed.
# If you have trouble, consider running an archive call individually without the Pool
# or using the bash script instead.
2025/09/12 01:49:54 UTC | [pid=2620] Starting build=sample
Building object sample from: ['yang']
Changing Look AGN- Yang et al: 31
Object sample size, after duplicates removal: 30
Object sample saved to: /home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/light_curves/output/lightcurves-demo-Yang-sample/object_sample.ecsv
2025/09/12 01:49:56 UTC
2025/09/12 01:49:56 UTC | [pid=2658] Starting build=lightcurves, archive=heasarc
2025/09/12 01:49:56 UTC | [pid=2659] Starting build=lightcurves, archive=icecube
2025/09/12 01:49:56 UTC | [pid=2656] Starting build=lightcurves, archive=gaia
2025/09/12 01:49:56 UTC | [pid=2660] Starting build=lightcurves, archive=tess_kepler
2025/09/12 01:49:56 UTC | [pid=2661] Starting build=lightcurves, archive=wise
2025/09/12 01:49:56 UTC | [pid=2657] Starting build=lightcurves, archive=hcv
working on mission
FERMIGTRIG
INFO
: Query finished. [astroquery.utils.tap.core]
working on mission
SAXGRBMGRB
Light curves saved to:
parquet_dir=/home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/light_curves/output/lightcurves-demo-Yang-sample/lightcurves.parquet
file=archive=gaia/part0.snappy.parquet
2025/09/12 01:49:59 UTC
Light curves saved to:
parquet_dir=/home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/light_curves/output/lightcurves-demo-Yang-sample/lightcurves.parquet
file=archive=heasarc/part0.snappy.parquet
2025/09/12 01:50:00 UTC
Light curves saved to:
parquet_dir=/home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/light_curves/output/lightcurves-demo-Yang-sample/lightcurves.parquet
file=archive=icecube/part0.snappy.parquet
2025/09/12 01:50:00 UTC
No light curve data was returned from hcv.
Light curves saved to:
parquet_dir=/home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/light_curves/output/lightcurves-demo-Yang-sample/lightcurves.parquet
file=archive=wise/part0.snappy.parquet
2025/09/12 01:51:45 UTC
No light curve data was returned from tess_kepler.
CPU times: user 578 ms, sys: 205 ms, total: 783 ms
Wall time: 3min 3s
/home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/.tox/py311-buildhtml/lib/python3.11/site-packages/lightkurve/prf/__init__.py:7: UserWarning: Warning: the tpfmodel submodule is not available without oktopus installed, which requires a current version of autograd. See #1452 for details.
warnings.warn(
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(0.28136, -0.09792)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(21.70042, -8.66336)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(29.99, 0.55301)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(120.9482, 42.97751)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(127.88438, 36.77141)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(132.49079, 27.79135)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(137.38344, 47.79187)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(144.37634, 26.04227)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(144.39779, 32.54717)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(150.84779, 35.41774)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(152.97079, 54.70177)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(166.09679, 63.71812)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(166.22988, 1.31573)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(167.60604, -0.05948)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(168.90237, 5.74715)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(169.62359, 32.06669)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(173.12143, 3.9581)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(177.66379, 36.54955)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(178.11453, 32.16649)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(194.81977, 55.25198)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(199.87816, 67.89873)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(209.07708, -1.25389)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(209.73261, 49.57057)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(221.97599, 28.55671)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(233.48329, 1.17494)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(236.37346, 25.19108)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(237.57182, 41.65067)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(238.24283, 27.62461)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(238.66777, 36.49777)>".
No data found for target "<SkyCoord (ICRS): (ra, dec) in deg
(354.01239, 0.29138)>".
The light curves are saved in the “parquet_dir” directory. Each archive call writes data to a parquet file in its own subdirectory. These can be loaded together as a single dataset:
# copy/paste the directory path from the output above, or ask the helper for it like this:
parquet_dir = helpers.scale_up.run(build="parquet_dir", **kwargs_dict)
df_lc = pd.read_parquet(parquet_dir)
df_lc.sample(10)
| flux | err | archive | ||||
|---|---|---|---|---|---|---|
| objectid | label | band | time | |||
| 15 | Yang 18 | W2 | 58253.730149 | 1.265453 | 0.016357 | wise |
| 14 | Yang 18 | W2 | 57534.435063 | 0.506390 | 0.014204 | wise |
| 12 | Yang 18 | W1 | 58792.864904 | 0.143566 | 0.005325 | wise |
| 9 | Yang 18 | IceCube | 54610.762312 | 3.760000 | 0.000000 | icecube |
| 20 | Yang 18 | G | 57151.012703 | 0.061910 | 0.000996 | gaia |
| 29 | Yang 18 | G | 56959.971168 | 0.119768 | 0.001406 | gaia |
| 22 | Yang 18 | W1 | 59025.612651 | 0.490864 | 0.006408 | wise |
| 21 | Yang 18 | IceCube | 57825.702011 | 4.000000 | 0.000000 | icecube |
| 16 | Yang 18 | W2 | 58974.086039 | 0.247982 | 0.016058 | wise |
| 19 | Yang 18 | W2 | 58459.752508 | 0.774738 | 0.016176 | wise |
Now we can make figures:
_ = create_figures(df_lc=MultiIndexDFObject(data=df_lc), show_nbr_figures=1, save_output=False)
Done
3. Keyword arguments and script flags#
This example shows the python kwargs_dict and bash script flag options in more detail.
3.1 Python kwargs_dict#
kwargs_dict is a dictionary containing all keyword arguments for the run. It can contain:
names and keyword arguments for any of the
get_*_samplefunctions.keyword arguments for any of the
*_get_lightcurvesfunctions.other keyword arguments used directly by the helper. These options and their defaults are shown below, further documented in the helper’s
runfunction.
# show kwargs_dict defaults
helpers.scale_up.DEFAULTS
{'run_id': 'my-run',
'get_samples': ['Yang'],
'consolidate_nearby_objects': True,
'overwrite_existing_sample': True,
'archives': ['Gaia',
'HCV',
'HEASARC',
'IceCube',
'PanSTARRS',
'TESS_Kepler',
'WISE',
'ZTF'],
'overwrite_existing_lightcurves': True,
'use_yaml': False,
'yaml_filename': 'kwargs.yml',
'sample_filename': 'object_sample.ecsv',
'parquet_dataset_name': 'lightcurves.parquet'}
# show parameter documentation
print(helpers.scale_up.run.__doc__)
Run the light_curve_collector step indicated by `build`.
Parameters
----------
build : str
Which step to run. Generally either "sample" or "lightcurves". Can also be either "kwargs"
or any of the `kwargs_dict` keys, and then the full set of kwargs is built from `kwargs_dict`
and either the whole dictionary is returned (if "kwargs") or the value of this key is returned.
kwargs_dict : dict
Key/value pairs for the build function. This can include any key in the dict
`helpers.scale_up.DEFAULTS` plus "archive".
run_id : str
ID for this run. This is used to name the output subdirectory ("base_dir") where the
scale_up helper will read/write files.
get_samples : list or dict[dict]
Names of get_<name>_sample functions from which to gather the object sample.
To send keyword arguments for any of the named functions, use a dict with key=name
value=dict of keyword arguments for the named function). Defaults will be
used for any parameter not provided.
consolidate_nearby_objects : bool
Whether to consolidate nearby objects in the sample. Passed to the clean_sample function.
overwrite_existing_sample : bool
Whether to overwrite an existing .ecsv file. If false and the file exists, the sample will simply
be loaded from the file and returned.
archive : str
Name of a <name>_get_lightcurves archive function to call when building light curves.
archives : list or dict[dict]
Names of <name>_get_lightcurves functions. Use a dict (key=name, value=dict of kwargs for the named
function) to override defaults for the named function. **Note that this does not determine which
archives are actually called** (unlike `get_samples` with sample names). This is because the
scale_up helper can only run one function at a time, so a single archive name must be passed separately
when building light curves.
overwrite_existing_lightcurves : bool
Whether to overwrite an existing .parquet file. If false and the file exists, the light curve
data will simply be loaded from the file and returned.
use_yaml : bool
Whether to load additional kwargs from a yaml file.
yaml_filename : str
Name of the yaml to read/write, relative to the base_dir.
sample_filename: str
Name of the `sample_table` .ecsv file to read/write, relative to the base_dir.
parquet_dataset_name : str
Name of the directory to read/write the parquet dataset, relative to the base_dir.
The dataset will contain one .parquet file for each archive that returned light curve data.
Returns
-------
Various
Depending on the `build` parameter, returns the result of the corresponding build function.
3.2 Bash script flags#
Use the -h (help) flag to view the script’s flag options:
# show flag documentation
!bash code_src/helpers/scale_up.sh -h
---- scale_up.sh ----
Use this script to launch and monitor a large-scale "run" to load a sample of target objects
from the literature and collect their multi-wavelength light curves from various archives.
For complete usage information, see the scale_up notebook tutorial.
FLAG OPTIONS
------------
Required flags:
-r 'run_id'
ID for the run. No spaces or special characters.
Determines the name of the output directory.
Can be used in multiple script calls to manage the same run.
Flags used to launch a run (optional):
-a 'archive names'
Space-separated list of archive names like 'HEASARC PanSTARRS WISE' (case insensitive),
or a shortcut, e.g., 'all'.
The get_<name>_lightcurves function will be called once for each name.
If this flag is not supplied, no light-curve data will be retrieved.
-d 'key=value'
Any top-level key/value pair in the python kwargs_dict where the
value is a basic type (e.g., bool or string, but not list or dict).
Repeat the flag to send multiple kwargs.
For more flexibility, use the -j flag and/or store the kwargs_dict as a
yaml file and use: -d 'use_yaml=true'. Order of precedence is dict, json, yaml.
-j 'json string'
The python kwargs_dict as a json string. An example usage is:
-j '{get_samples: {SDSS: {num: 50}}, archives: {ZTF: {nworkers: 8}}}'
The string can be created in python by first constructing the dictionary and then using:
>>> import json
>>> json.dumps(kwargs_dict)
Copy the output, including the surrounding single quotes ('), and paste it as the flag value.
Other flags (optional):
These must be used independently and cannot be combined with any other optional flag.
-t 'nsleep'
Use this flag to monitor top after launching a run. This will filter for PIDs
launched by 'run_id' and save the output to a log file once every 'nsleep' interval.
'nsleep' will be passed to the sleep command, so values like '10s' and '30m' are allowed.
The python helper can load the output.
This option is only available on Linux machines.
-k (kill)
Use this flag to kill all processes that were started using the given 'run_id'.
This option is only available on Linux machines.
-h (help)
Print this help message.
3.3 Using a yaml file#
It can be convenient to save the parameters in a yaml file, especially when using the bash script or in cases where you want to store parameters for later reference or re-use.
Define an extended set of parameters and save it as a yaml file:
yaml_run_id = "demo-kwargs-yaml"
get_samples = {
"green": {},
"ruan": {},
"papers_list": {
"paper_kwargs": [
{"paper_link": "2022ApJ...933...37W", "label": "Galex variable 22"},
{"paper_link": "2020ApJ...896...10B", "label": "Palomar variable 20"},
]
},
"SDSS": {"num": 10, "zmin": 0.5, "zmax": 2, "randomize_z": True},
"ZTF_objectid": {"objectids": ["ZTF18aabtxvd", "ZTF18aahqkbt", "ZTF18abxftqm", "ZTF18acaqdaa"]},
}
archives = {
"Gaia": {"search_radius": 2 / 3600},
"HEASARC": {"catalog_error_radii": {"FERMIGTRIG": 1.0, "SAXGRBMGRB": 3.0}},
"IceCube": {"icecube_select_topN": 4, "max_search_radius": 2.0},
"WISE": {"radius": 1.5, "bandlist": ["W1", "W2"]},
"ZTF": {"nworkers": 6, "match_radius": 2 / 3600},
}
yaml_kwargs_dict = {
"get_samples": get_samples,
"consolidate_nearby_objects": False,
"archives": archives,
}
helpers.scale_up.write_kwargs_to_yaml(run_id=yaml_run_id, **yaml_kwargs_dict)
kwargs written to /home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/light_curves/output/lightcurves-demo-kwargs-yaml/kwargs.yml
The path to the yaml file is printed in the output above.
You can alter the contents of the file as you like.
To use the file, set the kwarg use_yaml=True.
Python example for the get-sample step:
sample_table = helpers.scale_up.run(build="sample", run_id=yaml_run_id, use_yaml=True)
2025/09/12 01:52:58 UTC | [pid=2620] Starting build=sample
Building object sample from: ['sdss', 'ztf_objectid', 'green', 'papers_list', 'ruan']
SDSS Quasar: 10
number of ztf coords added by Objectname: 4
Changing Look AGN- Green et al 2022: 19
number of sources added from Galex variable 22 :48
number of sources added from Palomar variable 20 :20
Changing Look AGN- Ruan et al 2016: 3
Object sample size: 104
Object sample saved to: /home/runner/work/fornax-demo-notebooks/fornax-demo-notebooks/light_curves/output/lightcurves-demo-kwargs-yaml/object_sample.ecsv
2025/09/12 01:53:04 UTC
WARNING: OverflowError converting to IntType in column specObjID, reverting to String. [astropy.io.ascii.fastbasic]
Bash example:
$ yaml_run_id=demo-kwargs-yaml
$ bash code_src/helpers/scale_up.sh -r "$yaml_run_id" -d "use_yaml=true" -a "Gaia HEASARC IceCube PanSTARRS WISE ZTF"
Appendix: What to expect#
Challenges of large-scale runs#
Scaling up to large sample sizes brings new challenges. Even if things go smoothly, each function call may need to use a lot more resources like CPU, RAM, and bandwidth, and may take much longer to complete. Also, we’ll want to run some functions in parallel to save time overall, but that will mean they must compete with each other for resources.
These issues are complicated by the fact that different combinations of samples and archive calls can trigger different problems. Inefficiencies in any part of the process – our code, archive backends, etc. – which may have been negligible at small scale can balloon into significant hurdles.
Problems can manifest in different ways. For example, progress may slow to a crawl, or it may run smoothly for several hours and then crash suddenly. If the job is running in the background, print statements and error messages may get lost and never be displayed for the user if they are not redirected to a file.
Needs and wants for large-scale runs#
The main goal is to reduce the total time it takes to run the full code, so we want to look for opportunities to parallelize. We can group the light_curve_collector code into two main steps: (1) gather the target object sample; then (2) generate light curves by querying the archives and standardizing the returned data. All of the archive calls have to wait for the sample to be available before starting, but then they can run independently in parallel. This is fortunate, since gathering the sample does not take long compared to the archive calls.
It is useful to be able to monitor the run’s resource usage and capture print statements, error messages, etc. to log files in order to understand if/when something goes wrong. Even with parallelization, gathering light curves for a large sample of objects is likely to take a few hours at least. So we want to automate the monitoring tasks as much as possible.
If the run fails, we’d like to be able to restart it without having to redo steps that were previously successful.
To accomplish this, the inputs and outputs need to be less tightly coupled than they are in the light_curve_collector notebook.
Specifically, we want to save the sample and light curve data to file as soon as each piece is collected, and we want the archive functions to be able to get the sample_table input from file.
The python helper and bash script were specifically designed to fulfill many of these wants and needs.
Advice for the user#
Getting started:
Skim this notebook to understand the process, available options, and potential sticking points.
Try things with a small sample size first, then scale up to your desired full sample.
Don’t be surprised if something goes wrong. Every new combination of factors can present different challenges and reasons for the code to fail. This includes the sample selection, which archives are called and what parameters are used, runtime environment, machine CPU and RAM capabilities, network bandwidth, etc. Scaling up any code base comes with challenges, and some of these cannot be fully managed by pre-written code. You may need to observe how the code performs, diagnose a problem, and adapt the input parameters, machine size, etc. in order to successfully execute.
To execute a run:
Define all of the get-sample and get-lightcurve parameters.
Launch the run by calling the bash script or some other multi-processing method. Capturing stdout, stderr and
topoutput to log files is highly recommended.If a get-lightcurve (get-sample) job exits without writing a .parquet (.ecsv) file, inspect the logs and
topoutput to try to determine the reason it failed. It could be anything from a missing python library (install it), to an archive encountering an internal error (wait a bit and try again), to the job getting killed prematurely because its needs exceeded the available RAM (try a machine with more RAM, a smaller sample size, or running fewer archive calls at a time), to many other things.
About this notebook#
Authors: Troy Raen, Jessica Krick, Brigitta Sipőcz, Shoubaneh Hemmati, Andreas Faisst, David Shupe, and the Fornax team
Contact: For help with this notebook, please open a topic in the Fornax Community Forum “Support” category.
